Stepwise Regression
Stepwise Regression to Select Appropriate Models
stepwiselm creates a linear model and automatically adds to or trims the model. To create a small model, start from a constant model. To create a large model, start with a model containing many terms. A large model usually has lower error as measured by the fit to the original data, but might not have any advantage in predicting new data.
stepwiselm can use all the name-value options from fitlm, with additional options relating to the starting and bounding models. In particular:
For a small model, start with the default lower bounding model:
'constant'(a model that has no predictor terms).The default upper bounding model has linear terms and interaction terms (products of pairs of predictors). For an upper bounding model that also includes squared terms, set the
Uppername-value pair to'quadratic'.
Compare Large and Small Stepwise Models
This example shows how to compare models that stepwiselm returns starting from a constant model and starting from a full interaction model.
Load the carbig data and create a table from some of the data.
load carbig
tbl = table(Acceleration,Displacement,Horsepower,Weight,MPG);Create a mileage model stepwise starting from the constant model.
mdl1 = stepwiselm(tbl,'constant','ResponseVar','MPG')
1. Adding Weight, FStat = 888.8507, pValue = 2.9728e-103 2. Adding Horsepower, FStat = 3.8217, pValue = 0.00049608 3. Adding Horsepower:Weight, FStat = 64.8709, pValue = 9.93362e-15
mdl1 =
Linear regression model:
MPG ~ 1 + Horsepower*Weight
Estimated Coefficients:
Estimate SE tStat pValue
__________ __________ _______ __________
(Intercept) 63.558 2.3429 27.127 1.2343e-91
Horsepower -0.25084 0.027279 -9.1952 2.3226e-18
Weight -0.010772 0.00077381 -13.921 5.1372e-36
Horsepower:Weight 5.3554e-05 6.6491e-06 8.0542 9.9336e-15
Number of observations: 392, Error degrees of freedom: 388
Root Mean Squared Error: 3.93
R-squared: 0.748, Adjusted R-Squared: 0.746
F-statistic vs. constant model: 385, p-value = 7.26e-116
Create a mileage model stepwise starting from the full interaction model.
mdl2 = stepwiselm(tbl,'interactions','ResponseVar','MPG')
1. Removing Acceleration:Displacement, FStat = 0.024186, pValue = 0.8765 2. Removing Displacement:Weight, FStat = 0.33103, pValue = 0.56539 3. Removing Acceleration:Horsepower, FStat = 1.7334, pValue = 0.18876 4. Removing Acceleration:Weight, FStat = 0.93269, pValue = 0.33477 5. Removing Horsepower:Weight, FStat = 0.64486, pValue = 0.42245
mdl2 =
Linear regression model:
MPG ~ 1 + Acceleration + Weight + Displacement*Horsepower
Estimated Coefficients:
Estimate SE tStat pValue
__________ __________ _______ __________
(Intercept) 61.285 2.8052 21.847 1.8593e-69
Acceleration -0.34401 0.11862 -2.9 0.0039445
Displacement -0.081198 0.010071 -8.0623 9.5014e-15
Horsepower -0.24313 0.026068 -9.3265 8.6556e-19
Weight -0.0014367 0.00084041 -1.7095 0.088166
Displacement:Horsepower 0.00054236 5.7987e-05 9.3531 7.0527e-19
Number of observations: 392, Error degrees of freedom: 386
Root Mean Squared Error: 3.84
R-squared: 0.761, Adjusted R-Squared: 0.758
F-statistic vs. constant model: 246, p-value = 1.32e-117
Notice that:
mdl1has four coefficients (theEstimatecolumn), andmdl2has six coefficients.The adjusted R-squared of
mdl1is0.746, which is slightly less (worse) than that ofmdl2,0.758.
Create a mileage model stepwise with a full quadratic model as the upper bound, starting from the full quadratic model:
mdl3 = stepwiselm(tbl,'quadratic','ResponseVar','MPG','Upper','quadratic');
1. Removing Acceleration:Horsepower, FStat = 0.075209, pValue = 0.78405 2. Removing Acceleration:Weight, FStat = 0.072756, pValue = 0.78751 3. Removing Horsepower:Weight, FStat = 0.12569, pValue = 0.72314 4. Removing Weight^2, FStat = 1.194, pValue = 0.27521 5. Removing Displacement:Weight, FStat = 1.2839, pValue = 0.25789 6. Removing Displacement^2, FStat = 2.069, pValue = 0.15114 7. Removing Horsepower^2, FStat = 0.74063, pValue = 0.39
Compare the three model complexities by examining their formulas.
mdl1.Formula
ans = MPG ~ 1 + Horsepower*Weight
mdl2.Formula
ans = MPG ~ 1 + Acceleration + Weight + Displacement*Horsepower
mdl3.Formula
ans = MPG ~ 1 + Weight + Acceleration*Displacement + Displacement*Horsepower + Acceleration^2
The adjusted values improve slightly as the models become more complex:
RSquared = [mdl1.Rsquared.Adjusted, ...
mdl2.Rsquared.Adjusted, mdl3.Rsquared.Adjusted]RSquared = 1×3
0.7465 0.7580 0.7599
Compare residual plots of the three models.
subplot(3,1,1) plotResiduals(mdl1) subplot(3,1,2) plotResiduals(mdl2) subplot(3,1,3) plotResiduals(mdl3)
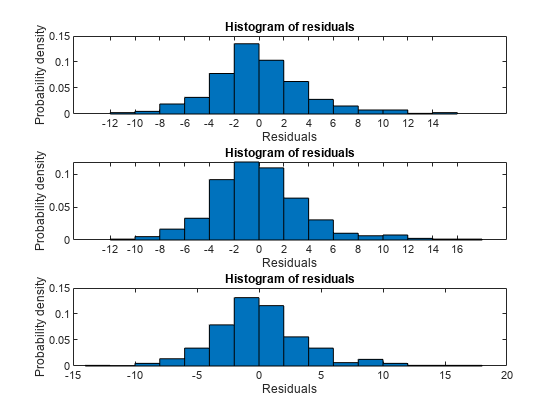
The models have similar residuals. It is not clear which fits the data better.
Interestingly, the more complex models have larger maximum deviations of the residuals:
Rrange1 = [min(mdl1.Residuals.Raw),max(mdl1.Residuals.Raw)]; Rrange2 = [min(mdl2.Residuals.Raw),max(mdl2.Residuals.Raw)]; Rrange3 = [min(mdl3.Residuals.Raw),max(mdl3.Residuals.Raw)]; Rranges = [Rrange1;Rrange2;Rrange3]
Rranges = 3×2
-10.7725 14.7314
-11.4407 16.7562
-12.2723 16.7927
See Also
fitlm | plotResiduals | stepwiselm | LinearModel
