cmdscale
Classical multidimensional scaling
Syntax
Y = cmdscale(D)
[Y,e] = cmdscale(D)
[Y,e] = cmdscale(D,p)
Description
Y = cmdscale(D) takes
an n-by-n distance matrix D,
and returns an n-by-p configuration
matrix Y. Rows of Y are
the coordinates of n points in p-dimensional
space for some p < n. When D is
a Euclidean distance matrix, the distances between those points are
given by D. p is the dimension
of the smallest space in which the n points whose
inter-point distances are given by D can be embedded.
[Y,e] = cmdscale(D) also
returns the eigenvalues of Y*Y'. When D is
Euclidean, the first p elements of e are
positive, the rest zero. If the first k elements
of e are much larger than the remaining (n-k),
then you can use the first k columns of Y as k-dimensional
points whose inter-point distances approximate D.
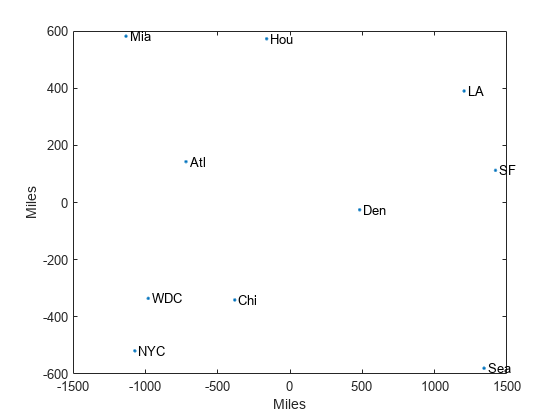
This can provide a useful dimension reduction for visualization, e.g.,
for k = 2.
D need not be a Euclidean distance matrix.
If it is non-Euclidean or a more general dissimilarity matrix, then
some elements of e are negative, and cmdscale chooses p as
the number of positive eigenvalues. In this case, the reduction to p or
fewer dimensions provides a reasonable approximation to D only
if the negative elements of e are small in magnitude.
[Y,e] = cmdscale(D,p) also accepts a positive integer
p between 1 and n. p
specifies the dimensionality of the desired embedding Y. If a
p dimensional embedding is possible, then Y
will be of size n-by-p and e
will be of size p-by-1. If only a q dimensional
embedding with q < p is possible, then Y will
be of size n-by-q and e will be
of size p-by-1. Specifying p may reduce the
computational burden when n is very large.
You can specify D as either a full dissimilarity
matrix, or in upper triangle vector form such as is output by pdist.
A full dissimilarity matrix must be real and symmetric, and have zeros
along the diagonal and positive elements everywhere else. A dissimilarity
matrix in upper triangle form must have real, positive entries. You
can also specify D as a full similarity matrix,
with ones along the diagonal and all other elements less than one. cmdscale transforms
a similarity matrix to a dissimilarity matrix in such a way that distances
between the points returned in Y equal or approximate sqrt(1-D).
To use a different transformation, you must transform the similarities
prior to calling cmdscale.
Examples
References
[1] Seber, G. A. F. Multivariate Observations. Hoboken, NJ: John Wiley & Sons, Inc., 1984.
Version History
Introduced before R2006a
See Also
mdscale | pdist | procrustes